|
InvivoGen
thp 1 lucia isg cells Thp 1 Lucia Isg Cells, supplied by InvivoGen, used in various techniques. Bioz Stars score: 94/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/thp 1 lucia isg cells/product/InvivoGen Average 94 stars, based on 1 article reviews
thp 1 lucia isg cells - by Bioz Stars,
2026-02
94/100 stars
|
Buy from Supplier |
|
InvivoGen
nf κb monocytes Nf κb Monocytes, supplied by InvivoGen, used in various techniques. Bioz Stars score: 96/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/nf κb monocytes/product/InvivoGen Average 96 stars, based on 1 article reviews
nf κb monocytes - by Bioz Stars,
2026-02
96/100 stars
|
Buy from Supplier |
|
InvivoGen
thp 1 dualtm Thp 1 Dualtm, supplied by InvivoGen, used in various techniques. Bioz Stars score: 96/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/thp 1 dualtm/product/InvivoGen Average 96 stars, based on 1 article reviews
thp 1 dualtm - by Bioz Stars,
2026-02
96/100 stars
|
Buy from Supplier |
|
InvivoGen
thp1 xblue md2 cd14 cell line Thp1 Xblue Md2 Cd14 Cell Line, supplied by InvivoGen, used in various techniques. Bioz Stars score: 95/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/thp1 xblue md2 cd14 cell line/product/InvivoGen Average 95 stars, based on 1 article reviews
thp1 xblue md2 cd14 cell line - by Bioz Stars,
2026-02
95/100 stars
|
Buy from Supplier |
|
InvivoGen
cells thp1 cells  Cells Thp1 Cells, supplied by InvivoGen, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/cells thp1 cells/product/InvivoGen Average 93 stars, based on 1 article reviews
cells thp1 cells - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
InvivoGen
asc speck reporter  Asc Speck Reporter, supplied by InvivoGen, used in various techniques. Bioz Stars score: 95/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/asc speck reporter/product/InvivoGen Average 95 stars, based on 1 article reviews
asc speck reporter - by Bioz Stars,
2026-02
95/100 stars
|
Buy from Supplier |
|
InvivoGen
asc deficient thp1 defasc cell line  Asc Deficient Thp1 Defasc Cell Line, supplied by InvivoGen, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/asc deficient thp1 defasc cell line/product/InvivoGen Average 93 stars, based on 1 article reviews
asc deficient thp1 defasc cell line - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
InvivoGen
thp 1 defcasp1  Thp 1 Defcasp1, supplied by InvivoGen, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/thp 1 defcasp1/product/InvivoGen Average 93 stars, based on 1 article reviews
thp 1 defcasp1 - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
InvivoGen
isg cells 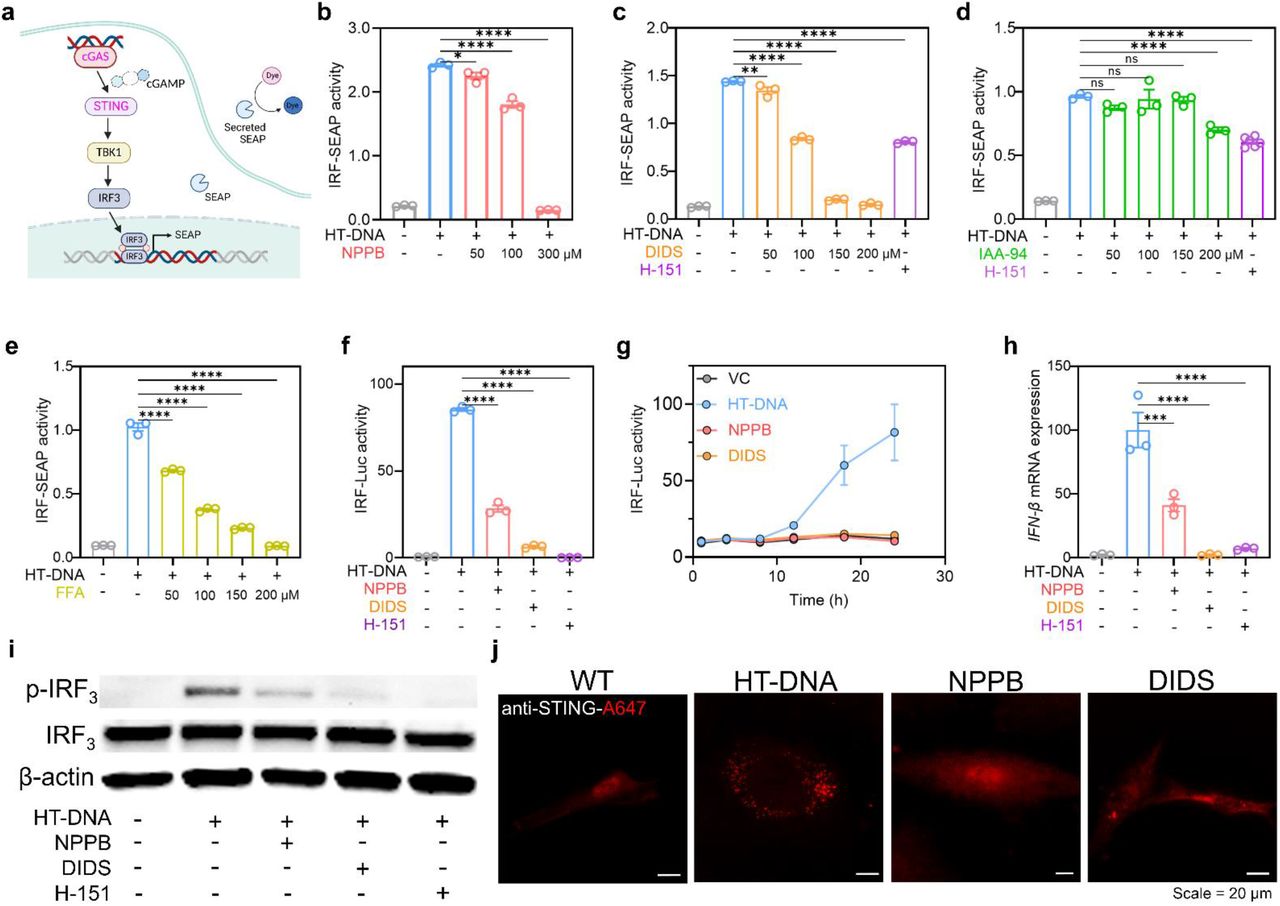 Isg Cells, supplied by InvivoGen, used in various techniques. Bioz Stars score: 95/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/isg cells/product/InvivoGen Average 95 stars, based on 1 article reviews
isg cells - by Bioz Stars,
2026-02
95/100 stars
|
Buy from Supplier |
|
InvivoGen
human monocyte cell line thp1 hmgb1 lucia  Human Monocyte Cell Line Thp1 Hmgb1 Lucia, supplied by InvivoGen, used in various techniques. Bioz Stars score: 93/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/human monocyte cell line thp1 hmgb1 lucia/product/InvivoGen Average 93 stars, based on 1 article reviews
human monocyte cell line thp1 hmgb1 lucia - by Bioz Stars,
2026-02
93/100 stars
|
Buy from Supplier |
|
InvivoGen
thp1 ki hsting r232 cells  Thp1 Ki Hsting R232 Cells, supplied by InvivoGen, used in various techniques. Bioz Stars score: 94/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/thp1 ki hsting r232 cells/product/InvivoGen Average 94 stars, based on 1 article reviews
thp1 ki hsting r232 cells - by Bioz Stars,
2026-02
94/100 stars
|
Buy from Supplier |
|
InvivoGen
monocytic cell line thp 1 lucia nf κb monocytes  Monocytic Cell Line Thp 1 Lucia Nf κb Monocytes, supplied by InvivoGen, used in various techniques. Bioz Stars score: 94/100, based on 1 PubMed citations. ZERO BIAS - scores, article reviews, protocol conditions and more https://www.bioz.com/result/monocytic cell line thp 1 lucia nf κb monocytes/product/InvivoGen Average 94 stars, based on 1 article reviews
monocytic cell line thp 1 lucia nf κb monocytes - by Bioz Stars,
2026-02
94/100 stars
|
Buy from Supplier |
Image Search Results

Journal: Scientific Reports
Article Title: Novel long non-coding RNAs associated with inflammation and macrophage activation in human
doi: 10.1038/s41598-023-30568-1
Figure Lengend Snippet: PMA induced differentiation of THP1 monocytes into macrophages (THP1-Mɸ) and their response to LPS-stimulation. ( A ) THP1 cells (monocytes) were differentiated using PMA (25 nM, 72 h) on a coverslip (35 mm cell culture plate). Cells were immuno-stained with CD68 antibody (mouse) followed by FITC-conjugated secondary antibodies, counterstained with DNA binding dye DAPI, mounted, and analyzed under a fluorescence microscope. Images taken at 40X resolution (bar = 50 μm). ( B ) LPS-stimulation of THP1 (monocytes) and THP1-Mɸ. THP1 and THP1-Mɸ cells were treated with LPS (1 μg/mL, 4 h) independently, total RNA was isolated, reverse transcribed to cDNA, and analyzed by RT-qPCR for expression of IL-6 and IL-1β. Each experiment was repeated at least thrice with three parallel replicates. β-Actin was used as loading control. Data represent mean ± SEM (n = 3); * p < 0.05, ** p < 0.001, *** p < 0.0001.
Article Snippet: 3 × 10 6
Techniques: Cell Culture, Staining, Binding Assay, Fluorescence, Microscopy, Isolation, Reverse Transcription, Quantitative RT-PCR, Expressing, Control

Journal: Scientific Reports
Article Title: Novel long non-coding RNAs associated with inflammation and macrophage activation in human
doi: 10.1038/s41598-023-30568-1
Figure Lengend Snippet: RNAseq analysis of LPS-treated macrophages. THP1 c ells were differentiated into PMA into macrophages (THP1-MΦ), treated with LPS (1.0 μg/mL) for 4 h. Total RNA was extracted from the control cells (C1–C3) and LPS-treated THP1-MΦ cells (L1–L3), quantified and subjected to ribo-depletion followed by library construction using the Illumina TruSeq Stranded Total RNA Library Prep Gold kit. Libraries were sequenced in an Illumina NovaSeq instrument. Differential gene expression analysis was done using the R package edgeR (v3.10.5) (PMID:19910308). Differentially expressed genes (log2 -old) were plotted as a heatmap. Cutoff values of absolute fold change greater than 2.0 and FDR ≤ 0.05 were used to select for differentially expressed genes between sample group comparisons.
Article Snippet: 3 × 10 6
Techniques: Control, Gene Expression

Journal: Scientific Reports
Article Title: Novel long non-coding RNAs associated with inflammation and macrophage activation in human
doi: 10.1038/s41598-023-30568-1
Figure Lengend Snippet: Pathways affected by LPS-stimulation of THP1-MΦ. RNAseq data was analysis using Panther-based data analysis to identify different signaling pathways that are affected by LPS-stimulation of macrophages.
Article Snippet: 3 × 10 6
Techniques: Protein-Protein interactions

Journal: Scientific Reports
Article Title: Novel long non-coding RNAs associated with inflammation and macrophage activation in human
doi: 10.1038/s41598-023-30568-1
Figure Lengend Snippet: LPS induces inflammation in THP1-macrophages (THP1-Mɸ). THP1-Mɸ cells were treated with LPS (1 μg/mL, 4 h), total RNA and proteins were isolated. RNA was reverse transcribed to cDNA and analyzed by RT-qPCR for expression of proinflammatory cytokines like IL-6 and IL-1β ( A ), as well as top upregulated protein coding genes (found in RNA-seq analysis) including ACOD1 and IDO1 at transcript level ( B ); and hLinfRNAs (1–5) ( C ). ( D ) Western blot analysis of protein coding genes. Proteins from the control and LPS-treated THP1-MΦ were analyzed by Western blot using antibodies against IL6, IL-1β, ACOD1, IDO1, and β-Actin (control). Bands were quantified and plotted in Fig. 4E. The specific region selected for each western blot is shown by red–rectangle in original respective western blot in the supplementary figure . Each experiment was repeated at least thrice with three parallel replicates. β-Actin was used as loading control. Data represent mean ± SEM (n = 3); * p < 0.05, ** p < 0.001, *** p < 0.0001.
Article Snippet: 3 × 10 6
Techniques: Isolation, Reverse Transcription, Quantitative RT-PCR, Expressing, RNA Sequencing, Western Blot, Control

Journal: Scientific Reports
Article Title: Novel long non-coding RNAs associated with inflammation and macrophage activation in human
doi: 10.1038/s41598-023-30568-1
Figure Lengend Snippet: hLinfRNAs are expressed in a dose-dependent manner in THP1-macrophages (THP1-Mɸ) under LPS induced inflammation. THP1-MΦ cells were treated with varying concentration of LPS (0.1- 1000 ng/mL, 4 h), total RNA was isolated and analyzed by RT-qPCR for expression of proinflammatory cytokines (IL6, IL-1β) and top 5 hLinfRNAs. β-Actin was used as loading control. Data represents mean ± SEM (n = 3).
Article Snippet: 3 × 10 6
Techniques: Concentration Assay, Isolation, Quantitative RT-PCR, Expressing, Control

Journal: Scientific Reports
Article Title: Novel long non-coding RNAs associated with inflammation and macrophage activation in human
doi: 10.1038/s41598-023-30568-1
Figure Lengend Snippet: Temporal expression of hLinfRNAs under LPS-stimulation of THP1-Mɸ. THP1-Mɸ cells were treated with LPS (1 μg/mL) for varying time periods. RNA was analyzed by RT-qPCR for expression of proinflammatory cytokines (IL6, IL-1β) and top 5 hLinfRNAs. Each experiment was repeated at least thrice with three parallel replicates. β-Actin was used as loading control. Data represents mean ± SEM (n = 3).
Article Snippet: 3 × 10 6
Techniques: Expressing, Quantitative RT-PCR, Control

Journal: Scientific Reports
Article Title: Novel long non-coding RNAs associated with inflammation and macrophage activation in human
doi: 10.1038/s41598-023-30568-1
Figure Lengend Snippet: hLinfRNAs are regulated by NF-κB signaling pathway in THP1-macrophages. THP1-MΦ cells were treated with IKKβί (SC-514, 25 μM, 1 h) followed by LPS (1 μg/mL). RNA and proteins were isolated from the control and LPS (with and without SC514) -treated cells and analyzed by RT-qPCR and Western blotting respectively. ( A-B ) Western blot analysis for the IκBα, phospho-IκBα, p65 and phospho-p65 (β-actin was used as a loading control). Quantifications are shown in panel 7B. The specific region selected for each western blot are shown by red–rectangle in the original respective western blots, supplementary figure . C-D) RT-qPCR analysis for the expression of pro-inflammatory cytokine (IL6, panel C ) and hLinfRNAs (1–5, panel D ). Data represents mean ± SEM (n = 3). * p < 0.05, ** p < 0.001, *** p < 0.0001.
Article Snippet: 3 × 10 6
Techniques: Isolation, Control, Quantitative RT-PCR, Western Blot, Expressing

Journal: Scientific Reports
Article Title: Novel long non-coding RNAs associated with inflammation and macrophage activation in human
doi: 10.1038/s41598-023-30568-1
Figure Lengend Snippet: Knockdown of hLinfRNA1 down-regulates the LPS-induced inflammatory response in macrophage. THP1-MΦ cells were transfected with hLinfRNA specific antisense oligonucleotide (ASO1 and ASO3) and scramble antisense for 48 h, stimulated with LPS (1 μg/mL) and incubated for additional 4 h. RNA was analyzed by RT-qPCR for expression of hLinfRNA1 and proinflammatory cytokines IL6, TNFα and IL1β (Fig. 8A) and PCR amplified product was analyzed in 2% agarose gel electrophoresis (Fig. 8B). The specific region selected for each agarose gel is shown by red–rectangle in the supplementary figure . Each experiment was repeated at least thrice with three parallel replicates. β-Actin was used as loading control. Data represents mean ± SEM (n = 3); * p < 0.05, ** p < 0.001, *** p < 0.0001.
Article Snippet: 3 × 10 6
Techniques: Knockdown, Transfection, Incubation, Quantitative RT-PCR, Expressing, Amplification, Agarose Gel Electrophoresis, Control

Journal: Clinical and Experimental Dental Research
Article Title: Hydrogen sulfide exposure induces NLRP3 inflammasome‐dependent IL‐1β and IL‐18 secretion in human mononuclear leukocytes in vitro
doi: 10.1002/cre2.69
Figure Lengend Snippet: The IL‐1ß and IL‐18 secretion of three THP1 cell lines exposed to 1 mM NaHS for 24 hr. Prior to NaHS, the cells were exposed to lipopolysaccharides. The THP1‐Null cells showed a statistically higher IL‐1ß and IL‐18 secretion when exposed to NaHS (Mann–Whitney U test, p = .0006 for IL‐1ß and p = .002 for IL‐18) compared with unexposed cells. When the other two cell lines were tested, both unable to form the NLRP3‐inflammasome, there was no difference in IL‐1ß and IL‐18 secretion when exposed to NaHS compared to control. The median of the group is shown as a vertical line
Article Snippet: In contrast, the
Techniques: MANN-WHITNEY

Journal: Cell Death Discovery
Article Title: Bacillus cereus cereolysin O induces pyroptosis in an undecapeptide-dependent manner
doi: 10.1038/s41420-024-01887-7
Figure Lengend Snippet: A THP-1 cells were incubated with or without (Ctrl) CLO at different doses for 1 h and then observed with a microscope. B The time-lapse images of THP-1 cells treated with CLO (100 nM). C The hemolytic activity of CLO at different doses was determined. D THP-1 cells were incubated with or without (Ctrl) CLO at different doses for 1 h, and then measured for LDH release. In panels A and B , arrows indicate membrane blebbing, and the scale bar is 30 μm. In panels C and D , values are shown as means ± SD ( N = 3). N, the number of replicates. ** p < 0.01(one-way ANOVA).
Article Snippet: THP-1-Null,
Techniques: Incubation, Microscopy, Activity Assay, Membrane

Journal: Cell Death Discovery
Article Title: Bacillus cereus cereolysin O induces pyroptosis in an undecapeptide-dependent manner
doi: 10.1038/s41420-024-01887-7
Figure Lengend Snippet: A , B THP-1 cells were treated with CLO (100 nM) or necroptosis inducer TBZ (TNFα, the SMAC mimetic BV-6 and Z-VAD) in the presence of DMSO or the inhibitors GSK’963, GSK’872, and GW806742X (targeting RIPK1, RIPK3, and MLKL respectively) for 1 h or 16 h. The cells were then subjected to LDH release determination ( A ) and microscopy after PI staining ( B ). Scale bar, 30 μm. C THP-1 cells were treated with or without (Ctrl) CLO (100 nM) in the presence of Q-VD-OPh, Ac-YVAD-CMK, Ac-DEVD-CMK, Z-LEVD-FMK, Z-IETD-FMK, or DMSO for 1 h. LDH release was then measured. D THP-1 cells were treated with or without (Ctrl) CLO (100 nM) in the presence or absence of different concentrations of Ac-YVAD-CMK for 1 h. LDH release was then measured. E , F THP-1 WT and THP-1 GSDMD-KO cells were treated with or without (Ctrl) CLO (100 nM) or nigericin (Nig) for 1 h. LDH ( E ) and IL-1β ( F ) release was then determined. G THP-1 GSDMD-KO cells were treated as C and then measured for LDH release. ** p < 0.01. NS no significance (one-way ANOVA test A , C , D , and G or student’s unpaired t test E and F . Values are shown as means ± SD ( N = 3). N the number of replicates.
Article Snippet: THP-1-Null,
Techniques: Microscopy, Staining

Journal: Cell Death Discovery
Article Title: Bacillus cereus cereolysin O induces pyroptosis in an undecapeptide-dependent manner
doi: 10.1038/s41420-024-01887-7
Figure Lengend Snippet: A J774A.1 cells were pretreated with MCC950, VX765, or DMSO for 1 h and then treated with CLO (CLO 100 nM) or ATP for 1 h. LDH release was then determined. B , C The cell lysate and supernatants from J774A.1 cells treated as above was immunoblotted with antibodies against Casp1, GSDMD, or β-actin (loading control). D , E PMA-differentiated THP-1 cells with or without (Null) deficiency in NLRP3 (NLRP3-KD) or Casp1 (Casp1-KD) were treated with or without (Ctrl) CLO (CLO 100 nM) or nigericin (Nig) for 1 h. LDH ( D ) and IL-1β ( E ) release was then determined. F The supernatant and the corresponding cell lysate from the above ( D , E ) treated J774A.1 cells were blotted with antibodies against Casp1, GSDMD, or β-actin (loading control). For panels A , D , and E , values are shown as means ± SD ( N = 3). N, the number of replicates. ** p < 0.01 (one-way ANOVA).
Article Snippet: THP-1-Null,
Techniques:

Journal: Cell Death Discovery
Article Title: Bacillus cereus cereolysin O induces pyroptosis in an undecapeptide-dependent manner
doi: 10.1038/s41420-024-01887-7
Figure Lengend Snippet: A , B J774A.1 cells pretreated with DCFH-DA were incubated with or without (Ctrl) DPI or NAC for 1 h. The cells were treated with or without CLO (10 nM) for 30 min. ROS production ( A ) and fluorescence intensity (λex, 488 nm; λem, 525 nm) ( B ) were then determined. C – G J774A.1 cells were pretreated with or without (−) DPI or NAC for 1 h and then treated with CLO (10 nM) or ATP for 1 h. Cell viability ( C ), LDH release ( D ), and IL-1β ( E ) release were determined. ASC speck (red) was detected by treating the cells with ASC-antibody and DAPI ( F ). The supernatant plus the corresponding cell lysate were blotted with antibody against Casp1, GSDMD, or β-actin (loading control) ( G ). H J774A.1 cells were incubated with or without (Ctrl) CLO (100 nM) or nigericin (Nig) for 30 min, and intracellular K + was then determined. I – K J774A.1 cells were pretreated with or without (−) different concentrations of KCl for 1 h, and then treated with or without (−) CLO (100 nM) for 1 h. LDH ( I ) and IL-1β release ( J ) was then determined. Immunoblot analysis of Casp-1, GSDMD, or β-actin was performed as above ( K ). Scale bars of panels A and F are 30 μm and 10 μm, respectively. For panels B – E and H – J , values are shown as means ± SD ( N = 3). N, the number of replicates. ** p < 0.01, * p < 0.05. NS no significance (one-way ANOVA).
Article Snippet: THP-1-Null,
Techniques: Incubation, Fluorescence, Western Blot

Journal: Cell Death Discovery
Article Title: Bacillus cereus cereolysin O induces pyroptosis in an undecapeptide-dependent manner
doi: 10.1038/s41420-024-01887-7
Figure Lengend Snippet: A CLO was incubated with a membrane lipid strip spotted with 15 lipids, and the bound CLO was detected by immunoblotting. B THP-1 cells were incubated with or without (Ctrl) CLO (100 nM) or cholesterol (Cho.)-pretreated CLO for 1 h. CLO was localized by immunofluorescence microscopy using dyLight 650 anti-6×His tag antibody. Scale bar, 30 μm. C – E J774A.1 cells were treated with or without (Ctrl) CLO (100 nM), nigericin (Nig), or Cho-pretreated CLO or Nig for 1 h. LDH ( C ) and IL-1β ( D ) release was then determined, and Casp1 and GSDMD cleavage was determined by Western blot with antibodies against Casp1, GSDMD, and β-actin (loading control) ( E ). For panels C and D , values are shown as means ± SD ( N = 3). N, the number of replicates. ** p < 0.01. NS no significance (student’s unpaired t test).
Article Snippet: THP-1-Null,
Techniques: Incubation, Membrane, Stripping Membranes, Western Blot, Immunofluorescence, Microscopy

Journal: Cell Death Discovery
Article Title: Bacillus cereus cereolysin O induces pyroptosis in an undecapeptide-dependent manner
doi: 10.1038/s41420-024-01887-7
Figure Lengend Snippet: A J774A.1 cells were treated with CLO (100 nM) or its mutants (100 nM) for 1 h, and LDH release was then determined. B Sterile defidrinated sheep blood was incubated with CLO (100 nM) or its mutants (100 nM) for 30 min and then detected for hemolysis. C A membrane lipid strip was incubated with the W477S-W479S mutant, and the bound protein was detected by immunoblotting. D THP-1 cells were incubated with or without (Ctrl) CLO (100 nM) or the W477S-W479S mutant (100 nM) for 1 h. The cells were stained with DAPI and subjected to immunofluorescence microscopy with dyLight 650 anti-6×His tag antibody. Scale bar, 30 μm. E , F J774A.1 cells were treated with or without (Ctrl) ATP, CLO (100 nM), or the W477S-W479S mutant (100 nM) for 1 h. The cells were determined for IL-1β release ( E ) and Casp1/GSDMD cleavage by immunoblot using antibodies against Casp1, GSDMD, and β-actin (loading control) ( F ). J774A.1 cells were treated with mutants (100 nM) for 1 h. LDH ( G ), IL-1β ( H ) and immunoblot analysis of Casp-1 and GSDMD ( I ) were assessed as above. ** p < 0.01. NS no significance (one-way ANOVA test A , B , G , and H or student’s unpaired t test E . Values are shown as means ± SD ( N = 3). N the number of replicates.
Article Snippet: THP-1-Null,
Techniques: Sterility, Incubation, Membrane, Stripping Membranes, Mutagenesis, Western Blot, Staining, Immunofluorescence, Microscopy

Journal: bioRxiv
Article Title: Chloride Homeostasis Regulates cGAS-STING Signaling
doi: 10.1101/2024.04.08.588475
Figure Lengend Snippet: ( a ) Schematic diagram illustrating the mechanism for monitoring cGAS-STING pathway activation via IRF3 reporter cells. Activation of IRF3 induces the production of secreted reporter enzyme. The reporter enzyme is secreted into cell culture supernatant and IRF 3 activation can be determined by reporter activity using ( b − e ) colorimetric (OD at 655nm) or ( f − g ) luminescent methods. ( b-e ) THP1-Blue ISG cells were pretreated with ( b ) 50-300 μM NPPB, ( c ) 50-200 μM DIDS, ( d ) 50-200 μM IAA-94, ( e ) 50-200 μM FFA, and 2.5 μM H-151 (STING inhibitor) for 1 h then transfected with 2×10 −2 μg/µL HT-DNA overnight. Error bars indicate the mean ± standard error of the mean (s.e.m.) of three independent measurements. * P < 0.05; ** P < 0.01; **** P < 0.0001. One-way analysis of variance (ANOVA) followed by Dunnett’s test for multiple comparison. ns, not significant. ( f-g ) RAW-Dual cells were pretreated with 100 μM NPPB, 150 μM DIDS, and 15 μM H-151 for 1 h then transfected with 2×10 −2 μg/µL HT-DNA for ( f ) overnight, ( g )1-24 h. Error bars indicate the mean ± standard error of the mean (s.e.m.) of three independent measurements. **** P < 0.0001. One-way analysis of variance (ANOVA) followed by Dunnett’s test for multiple comparison. (h) IFN-β mRNA expression levels in THP1 cells pretreated with 100 μM NPPB,150 μM DIDS,15μM H-151 for overnight and then transfected with 2×10 −2 µg/µL HT-DNA overnight. mRNA levels were measured by the RT-qPCR. Error bars indicate the mean ± standard error of the mean (s.e.m.) of three independent measurements. *** P < 0.001; **** P < 0.0001. One-way analysis of variance (ANOVA) followed by Dunnett’s test for multiple comparison. (i) Western blotting to measure the protein expression level of p-IRF 3 , IRF 3 and β-actin in THP-1 cells that were pretreated with 100 μM NPPB,150 μM DIDS, and 15 μM H-151 overnight and then transfected with 2×10 −2 µg/µL HT-DNA overnight. Experiments were performed in three biological replicates. (j) Primary human dermal fibroblasts (HDF) were pretreated with 100 μM NPPB, 150 μM DIDS for 18 h and then transfected with 2×10 −2 µg/µL HT-DNA for 6 h. Cells were fixed using paraformaldehyde and stained with anti-STING antibodies. Experiments were performed in three biological replicates.
Article Snippet: THP1-Blue™
Techniques: Activation Assay, Cell Culture, Activity Assay, Transfection, Comparison, Expressing, Quantitative RT-PCR, Western Blot, Staining

Journal: bioRxiv
Article Title: Chloride Homeostasis Regulates cGAS-STING Signaling
doi: 10.1101/2024.04.08.588475
Figure Lengend Snippet: (a) RAW-Dual cells were pretreated with 100 µM 2,3-cGAMP for 30 min, followed by washing with PBS, and then incubated with 100 μM NPPB and 15 μM H-151 overnight. (b) RAW-Dual cells were pretreated with 100 µM DMXAA or 100 µM 2,3-cGAMP for 30 min, followed by washing with PBS, and then incubated with 150 μM DIDS, and 15 μM H-151 overnight. (c) THP1-Blue ISG cells were pretreated with 100 μM NPPB, 200 µM IAA-94, 100 μM FFA, and 15 μM H-151 for 1 h and then stimulated with 30 µM MSA-2 overnight. (d) IFN-β mRNA expression levels were measured in THP1 cells pretreated with 100 μM NPPB, and 150 μM DIDS overnight and stimulated with 100 µM 2,3-cGAMP overnight. mRNA levels were measured using the RT-qPCR. (e) IFN-β mRNA expression levels were measured in THP1 cells pretreated with 100 μM NPPB, 150 μM DIDS and 15 μM H-151 overnight and stimulated with 30 µM MSA-2 overnight. mRNA levels were measured using the RT-qPCR. (a-e) Error bars indicate the mean ± standard error of the mean (s.e.m.) of three independent measurements. **** P < 0.0001. One-way analysis of variance (ANOVA) followed by Dunnett’s test for multiple comparison. ns, not significant. (f-g) Western blotting to measure the protein expression level of p-IRF 3 , IRF 3 and β-actin in THP-1 cells that were pretreated with ( f ) 100 μM NPPB, ( g ) 150 μM DIDS, and 15 μM H-151 overnight and then stimulated with 100 µM 2,3-cGAMP overnight. Experiments were performed in three biological replicates. (h) Immunofluorescent analysis of HDF cells pretreated with 100 μM NPPB, and 150 μM DIDS overnight and then stimulated with 30 µM MSA-2 for 6 h. (i) Percentage of cells with STING puncta correlating to ( h ). Experiments were performed in three biological replicates (>125 cells per trial). Error bars indicate the mean ± standard error of the mean (s.e.m.) of three independent measurements. **** P < 0.0001. One-way analysis of variance (ANOVA) followed by Dunnett’s test for multiple comparison.
Article Snippet: THP1-Blue™
Techniques: Incubation, Expressing, Quantitative RT-PCR, Comparison, Western Blot

Journal: Communications Biology
Article Title: Lysoptosis is an evolutionarily conserved cell death pathway moderated by intracellular serpins
doi: 10.1038/s42003-021-02953-x
Figure Lengend Snippet: a HT3 B3-WT (blue) or HT3 B3-KO (red) cells were subjected to nucleofection (nuc, +) in the absence (−) or presence (+) of 5 ug/ml LPS-EK (LPS), caspase-1 inhibitor, VX765, or lysosomal cysteine protease inhibitor, E64. An aliquot of cells were also not nucleofected (nuc, −). After 3 h, cells were stained with SG and Hoescht 33342 and imaged. Quantification: (# Sytox positive nuclei/# of blue nuclei) × 100. Means ± SD were compared using a two-tailed t -test (* P < 0.05). b Immunoblot of gasdermin D (GSDMD) and gasdermin E (GSDME) cleavage in THP1 cells treated with α-hemolysin (HlA); positive control. HT3 B3-WT /HT3 B3-KO were nucleofected with 0, 1, or 5 µg of LPS and left to recover for 1 h. Open arrowheads: full-length GSDMD and GSDME based on molecular mass. Black arrowheads: cleaved GSDMD. Dashed box: area contrast enhanced for clarity. Note, the positive control for GSDME cleavage is provided in Fig. . c , d Flow cytometry analysis of HT3 B3-WT /HT3 B3-KO cells treated with a lysosomotropic agent, LLOMe ( c ) or 1 µg/ml LPS ( d ). Cells were stained with the lysosomotropic dye, acridine orange (Y-axis), and the cell viability dye, Sytox™ blue (X-axis). Lines indicate the threshold fluorescence levels (gate) to determine each quadrant. Numbers indicate cell percentage in each quadrant. e Schematic representation of each quadrant. Quadrant 1 (Q1) contained live cells positive for lysosomal staining (acridine orange positive, Sytox blue negative). Quadrant 2 (Q2) contained dead cells with positive lysosomal staining (acridine orange positive, Sytox blue positive). Quadrant 3 (Q3) contained dead cells negative for lysosomal staining (acridine orange negative, Sytox blue positive). Quadrant 4 (Q4) contained live cells negative for lysosomal staining (acridine orange negative, Sytox blue negative). Arrows indicate the timing of fluorescence loss for different cell death pathways. If the lysosomal loss occurred prior to plasma membrane permeabilization, then the percentage of cells will increase from Q1 to Q4 to Q3 over time. If the lysosomal loss occurred after plasma membrane loss, then the percentage of cells will increase from Q1 to Q2 to Q3 over time. f , g Graphical representation of the average of three separate experiments indicating the percentage of cells treated with LLOMe ( f ) or nucleofected LPS ( g ) in HT3 B3-WT (blue) and HT3 B3-KO (red) cells. Error bars represent means ± SD. Uncropped immunoblots can be found in Supplementary Fig. .
Article Snippet: The
Techniques: Protease Inhibitor, Staining, Two Tailed Test, Western Blot, Positive Control, Flow Cytometry, Fluorescence, Membrane